Our lab uses various single-molecule techniques such as flow-stretching bead assay, optical tweezers, single-molecule FRET, single-molecule fluorescence microscopy, or their combinations to study various biological problems. We have also developed novel single-molecule techniques.
Flow-stretching Bead Assay
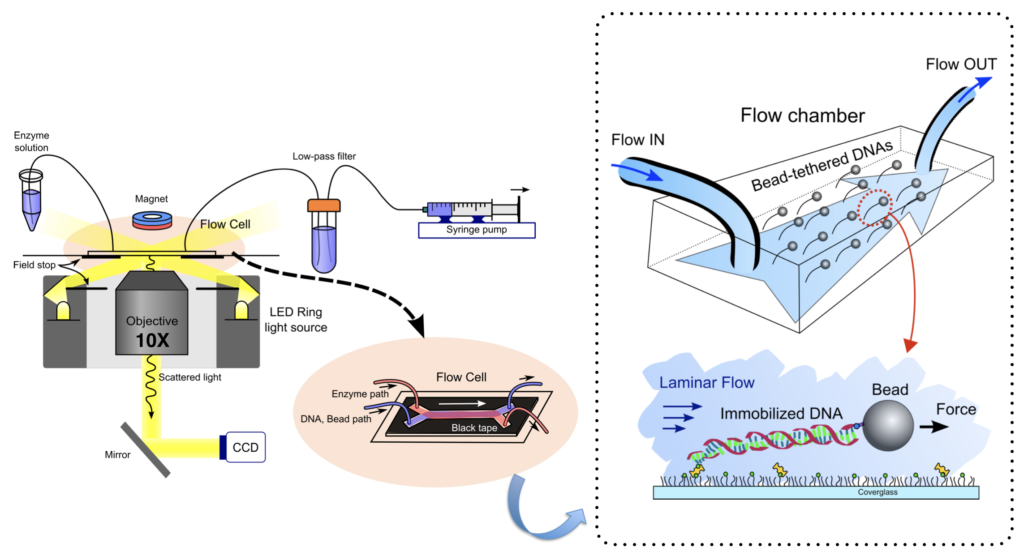
Single-molecule flow-stretching bead assay uses a well-defined drag force onto a micron-size super paramagnetic bead by regulated laminar flow. We monitor the position of the bead that is attached to the immobilized DNA to the surface to visualize the enzymatic activities on the DNA.
A cover glass is functionalized with PEG-biotin and PEG (with a mass ratio of 1:100, Laysan Bio) to minimize the nonspecific binding of DNA substrates or proteins. The flow chamber with 25.0 X 3.0 X 0.1 mm dimension is built up with a slide glass washed by Acetone and the functionalized cover glass. DNA substrates are attached to the cover glass surface of the flow chamber via a biotin-streptavidin linkage by flowing a DNA (~0.5 pM) in the blocking buffer for 10 min using a syringe pump (0.04 ml/min, Harvard apparatus). Anti-digoxigenin (anti-digoxigenin Fab, Roche) coated super-paramagnetic beads (2.8 um in diameter, Invitrogen) are introduced to the flow chamber where DNA substrates are immobilized, which is linked to the digoxigenin-end of the DNA. A hydrodynamic force by laminar flow of the buffer is applied to a tethered bead at ~2.2 pN. The magnetic force generated by a ring-shaped rare earth magnet is also applied to a bead at ~ 1 pN for avoiding a nonspecific interaction between the bead and the surface. The net force acting on the bead was 2.5 pN. The bead linked to DNA is imaged using an optical microscope under a 10 x objective (N.A. = 0.40, Olympus).
Publication: Yongmoon Jeon et al. PNAS (2016)
Correlative optical tweezers-line scanning confocal microscopy with extended depth of field
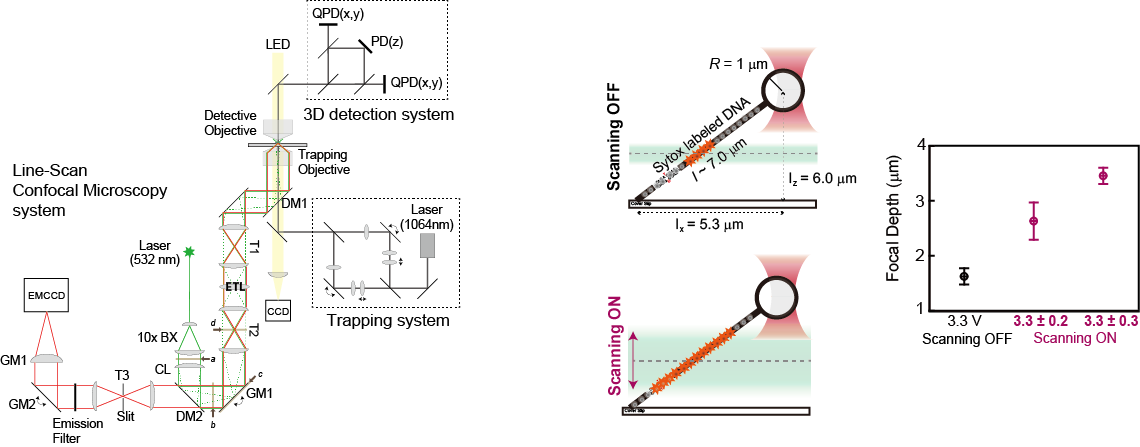
Real-time optical imaging combined with single-molecule manipulation broadens the horizons for acquiring information about the spatiotemporal localization and the mechanical details of target molecules. To obtain an optical signal outside the focal plane without unintended interruption of the force signal in single-molecule optical imaging-force spectroscopy, we developed an optical method to extend the depth of field in a high numerical aperture objective (> 1.2), required to visualize a single fluorophore. By axial scanning, using an electrically tunable lens with a fixed sample, we were successfully able to visualize the epidermal growth factor receptor (EGFR) moving along the three- dimensionally elongated filamentous actin bundles connecting cells (intercellular nanotube), while another EGFR on the intercellular nanotube was trapped by optical tweezers in living cells. Our approach is simple, fast and inexpensive, but it is powerful for imaging target molecules axially in single-molecule optical imaging-force spectroscopy.
Publication: Minhyeok Chang et al. Optics Express (2017)
Single-molecule polarization microscopy
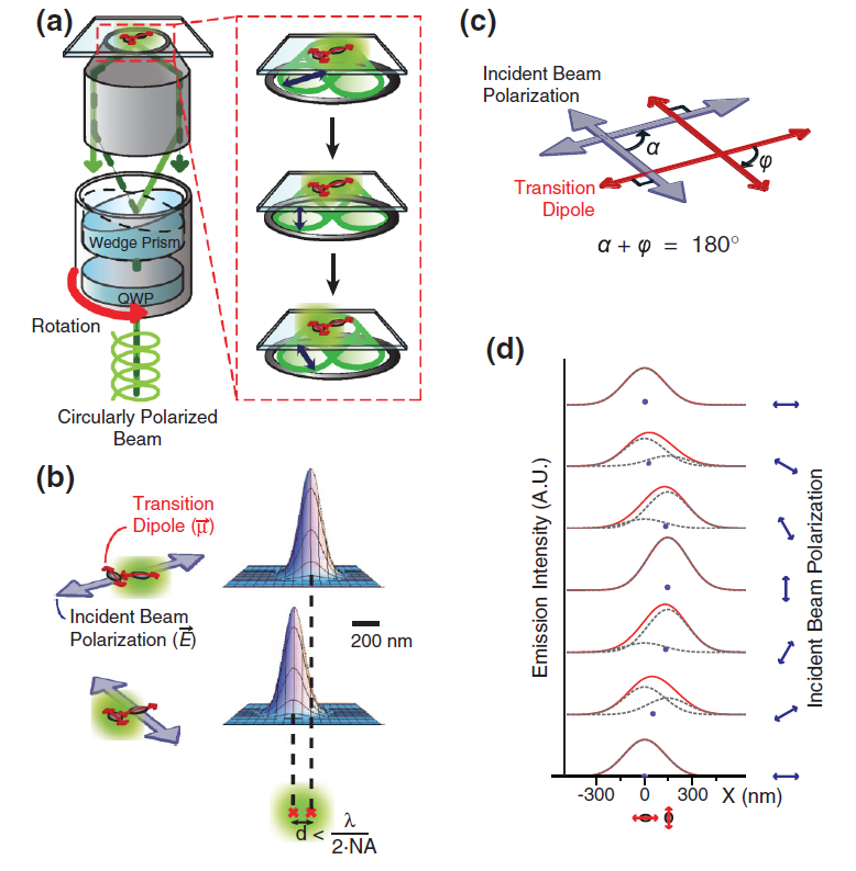
Modulation of fluorescence emission by linearly polarized excitation light can allow us to resolve spatially two fluorescent molecules within a diffraction limit and to determine simultaneously their precise dipole directions. Using polarization-dependent photoswitching, we imaged the 2D geometry of the DNA Holliday junction in a 10-nm length scale by measuring both the distance and the in-plane dipole angle between Cy3 emitters stacked onto the ends of two adjacent branches of the Holliday junction. The proposed polarization-modulated imaging technique provides a simple and nonstochastic imaging process to visualize the nanostructure, including directional information, of biomolecules beyond the diffraction limit.
Publication: Seongsil Lee et al. Physical Review Letter (2012)
