Intercellular Nanotubes for Long Distance Communication between Cells
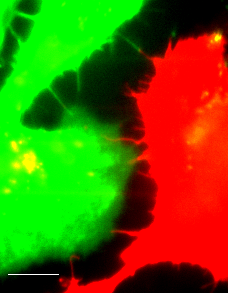
Fluorescent image (Line-scanning confocal microscopy) of actin filament labeled HeLa cells with different colors (Lifeact-GFP and -mCherry).
Scale bar = 10 μm
- In recent decades, intercellular nanotubes that connect cells over long distances of up to several cell diameters have been recognized as a new pathway for the communication between distant cells. However, it is not yet revealed how this curious structure extends over several hundred micrometers and remains robust for several hours.
- Using real-time imaging in live cells and dSTORM imaging in fixed cells, we revealed that the spatiotemporal processes of intercellular nanotube formation evolved from the fine nanostructure of filopodial bridges (two-filopodia complex, FB), which clearly demonstrates the mechanisms that underlie the formation and maintenance of actin-driven intercellular nanotubes. Surprisingly, we have observed helical structures as an intermediate state of intercellular nanotubes using super resolution microscopy and visualized the unique structure triggers the transition of the intermediate state to a single filopodium bridge that contacts to the paired cell body via cadherin-cadherin adhesion.
- Study on the role of FB twisting on its formation by force manipulation using optical tweezers and confocal microscopy revealed that FBs had a highly heterogeneous stiffness in a broad range in contrast to INTs. In a collaboration with theoretical biophysics lab of Jae-Hyung Jeon , our model of twisted FBs with a different number of turns explains this heterogeneity accurately. We expect that the accumulation of the torsional energy on FBs over the critical level can lead the transition from FBs to INTs by releasing one filopodium weakly attached to the other cell body.
Publication: Minhyeok Chang et al. BioRxiv (2018)
Translation initiation in cells
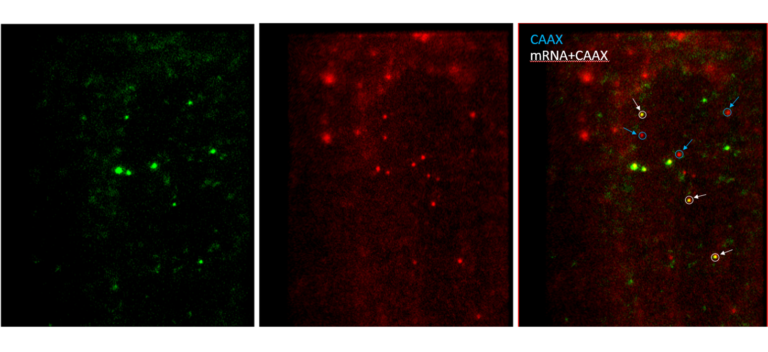
Fluorescent image (Line-scanning confocal microscopy) of mRNA being translated and CAAX labeled with different colors (scFv-GFP, CAAX-mCherry).
White circles on the right image indicate that translating mRNAs tethered with plasma membrane via CAAX.
- Translation that takes place in the cell is the process of converting the genetic information into protein and has three steps initiation, elongation, and termination. Live cell imaging has been evolved for decades, but single-molecule imaging of translation has not been achieved until recently due to weak signals of targets compared to background fluorescence and biomolecule’s thermal diffusion. In fact, the weak signals can be solved in principle by multiple fusion of fluorescent proteins to the target molecule. However, since many proteins are involved in the translation process, the multiple fusion of the fluorescent proteins to the target protein is far from natural cell behavior.
- To overcome the difficulties in single-molecule imaging, a novel technique has been developed recently (Yan et al. Cell 165, 976, 2016). The mRNA that PP7 stem loops are inserted into the 3’UTR region is tethered to the plasma membrane of the cell through the interaction between PP7 coated proteins and CAAX. The translation of the mRNA is monitored in real time using the SunTag system in the reporter mRNA. Furthermore, using membrane-permeable JF dyes, we can tag an organic dye easily into the protein of interest. Using TIRF microscopy, we visualize the translation initiation factors bound to mRNA and its translation processes with relatively high SNR . We will present the dynamic processes of the single-molecule 5’ cap-dependent translation in living cells.
